|
|
Wanderlust |
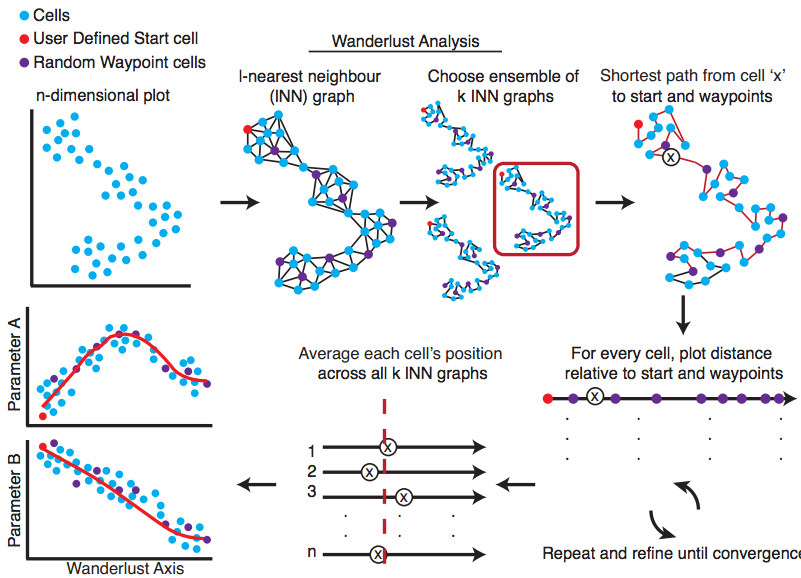 Wanderlust, a graph-based trajectory detection algorithm that receives multiparameter single-cell events as input and maps them onto a one-dimensional developmental trajectory. Cells are ordered along a trajectory that represents their most likely placement along a developmental continuum. Wanderlust, a graph-based trajectory detection algorithm that receives multiparameter single-cell events as input and maps them onto a one-dimensional developmental trajectory. Cells are ordered along a trajectory that represents their most likely placement along a developmental continuum.
|
If you use Wanderlust or its associated tool, CYT, for your publication, please cite the following article:
Bendall S.C., Davis K.L., Amir E.D, Tadmor M.D., Simonds E.F., Tiffany
J.C., Shenfeld D.K., Nolan G.P., Pe'er D. Single-Cell Trajectory Detection Uncovers Progression and Regulatory Coordination in Human
B cell Development, Cell. 2014.
|
You can download the data included in the paper here.
To generate and visualize wanderlust trajectories, download the associated matlab based tool, CYT, here. These instructions explain how to run wanderlust from within CYT. |
To use Wishbone (which aligns single cells from differentiation systems with bifurcating branches) please visit the Wishbone page. The software for Wishbone can be obtained here.
|
