|
|
DREVI and DREMI |
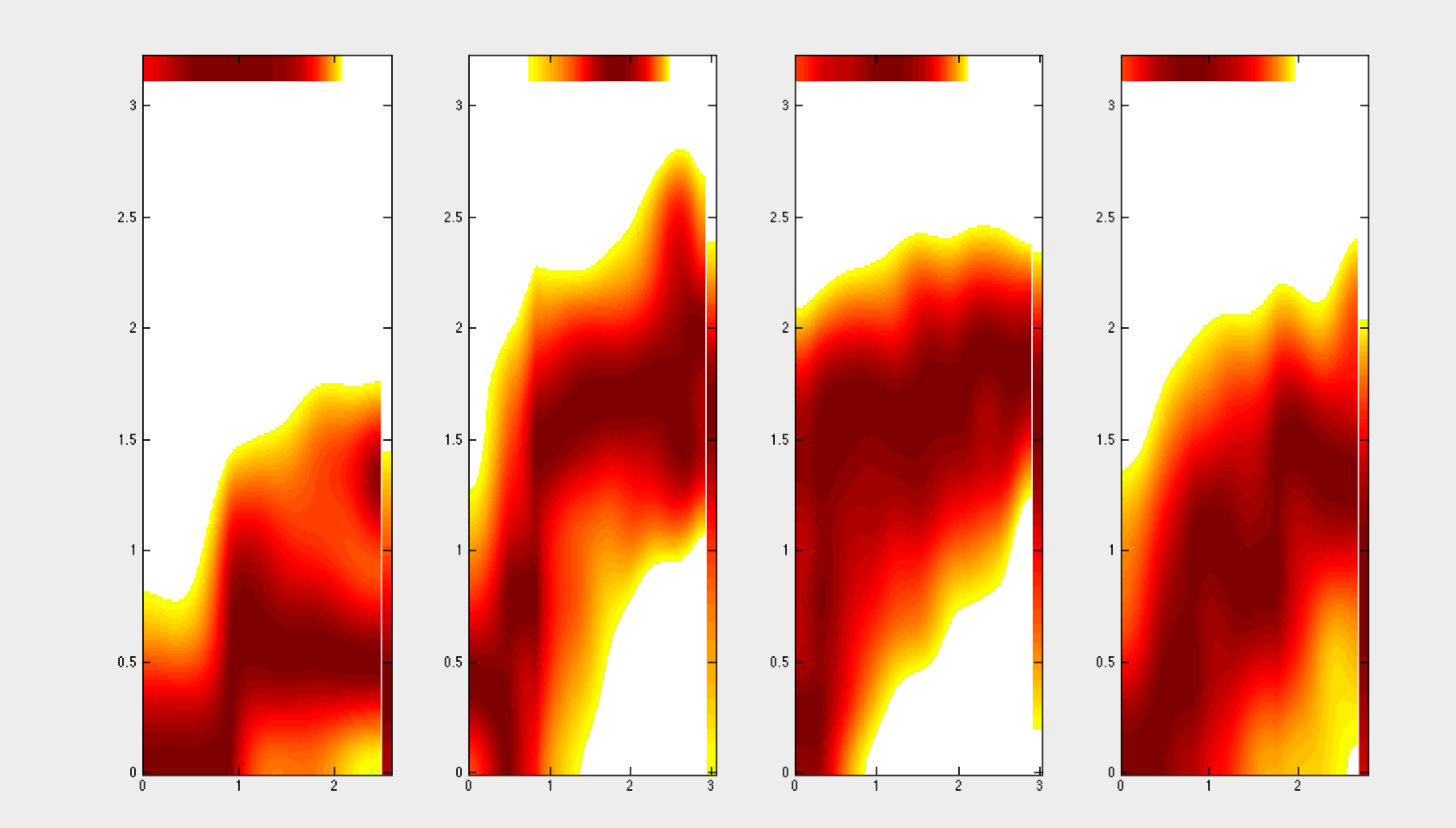 DREVI (conditional-Density Rescaled Visualization) and DREMI (conditional-Density Resampled estimate of Mutual Information) are algorithms for visualizing and scoring the strengths of pairwise interactions in single-cell data. Both algorithms
use conditional density estimation, where the distribution of the Y-variable is
conditioned on values of the the X-variable. In addition, this software can be used to extract information about the edge-response function (computed through regression) and also areas under the conditional-mean curve. DREVI (conditional-Density Rescaled Visualization) and DREMI (conditional-Density Resampled estimate of Mutual Information) are algorithms for visualizing and scoring the strengths of pairwise interactions in single-cell data. Both algorithms
use conditional density estimation, where the distribution of the Y-variable is
conditioned on values of the the X-variable. In addition, this software can be used to extract information about the edge-response function (computed through regression) and also areas under the conditional-mean curve. |
If you use this software for your publication, please cite the following article:
S. Krishnaswamy, M. Spitzer, M. Mingueneau, S.C. Bendall, E. L. Stone, O. Litvin, D. Pe'er, G. P. Nolan, Conditional Density-based Analysis of T cell Signaling in Single Cell Data, Science 2014.
|
You can download the data included in the paper here.
Download the matlab-based tool simpledremi for exploring DREVI and DREMI here. These instructions explain how to run simpledremi from matlab. |
|
